Our lab combines development of novel high-speed, 3D and multi-spectral optical imaging and microscopy systems with research into brain and tissue physiology. This page provides information about some of the systems that we have developed, and resources for building your own versions of our systems.
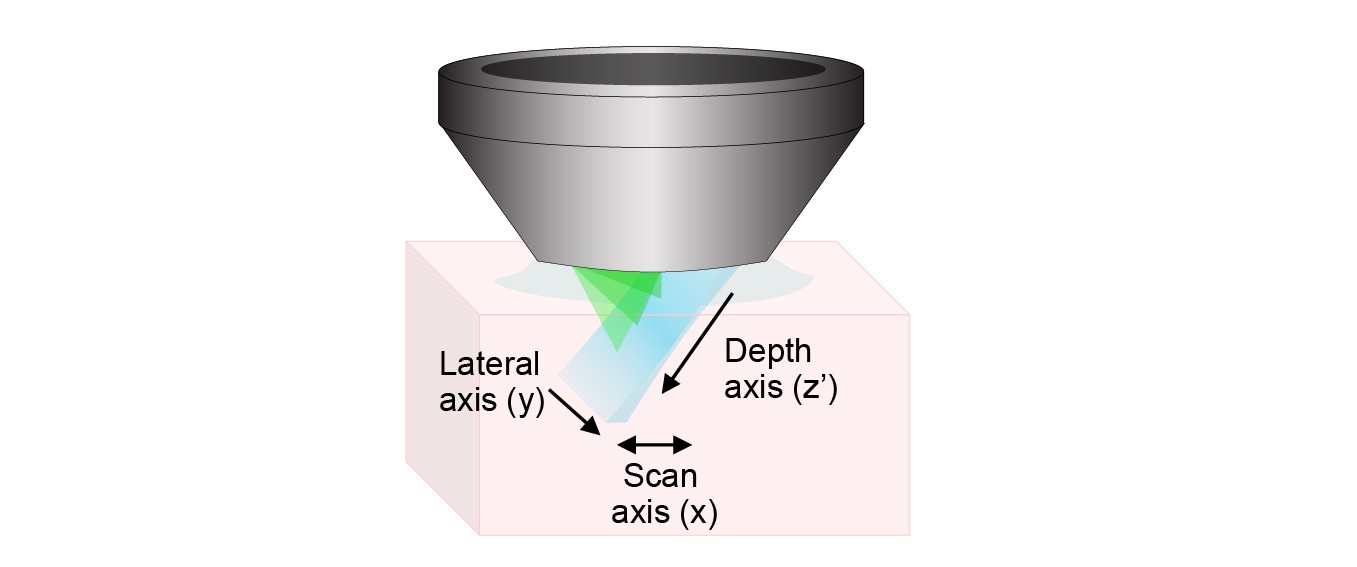
Swept, Confocally-Aligned Planar Excitation (SCAPE) microscopy is a technique for high-speed 3D microscopy in living organisms. SCAPE can image intact living samples including the intact mouse brain and freely moving organisms such as Drosophila melanogaster larvae and the zebrafish heart at up to 300 volumes per second through a single, stationary objective lens. SCAPE is licensed to Leica Microsystems for commercial development.
We originally introduced SCAPE in 2015, published in Nature Photonics. Our new paper in Nature Methods describes and demonstrates SCAPE 2.0, our new and improved, even faster, even higher resolution version of the system.
Our lab is working on many aspects of SCAPE, including developing more systems that can reach wider applications, including higher resolution, larger fields of view, and deeper imaging into scattering tissues using two-photon excitation. We are also working with a wide range of collaborators to apply SCAPE to specific research applications. For details on applications and collaborations see HERE.
We are also sharing SCAPE - helping labs around build their own versions of the system to accelerate their biomedical research. If you are interested in building a SCAPE microscope, please contact us for more details: [email protected]
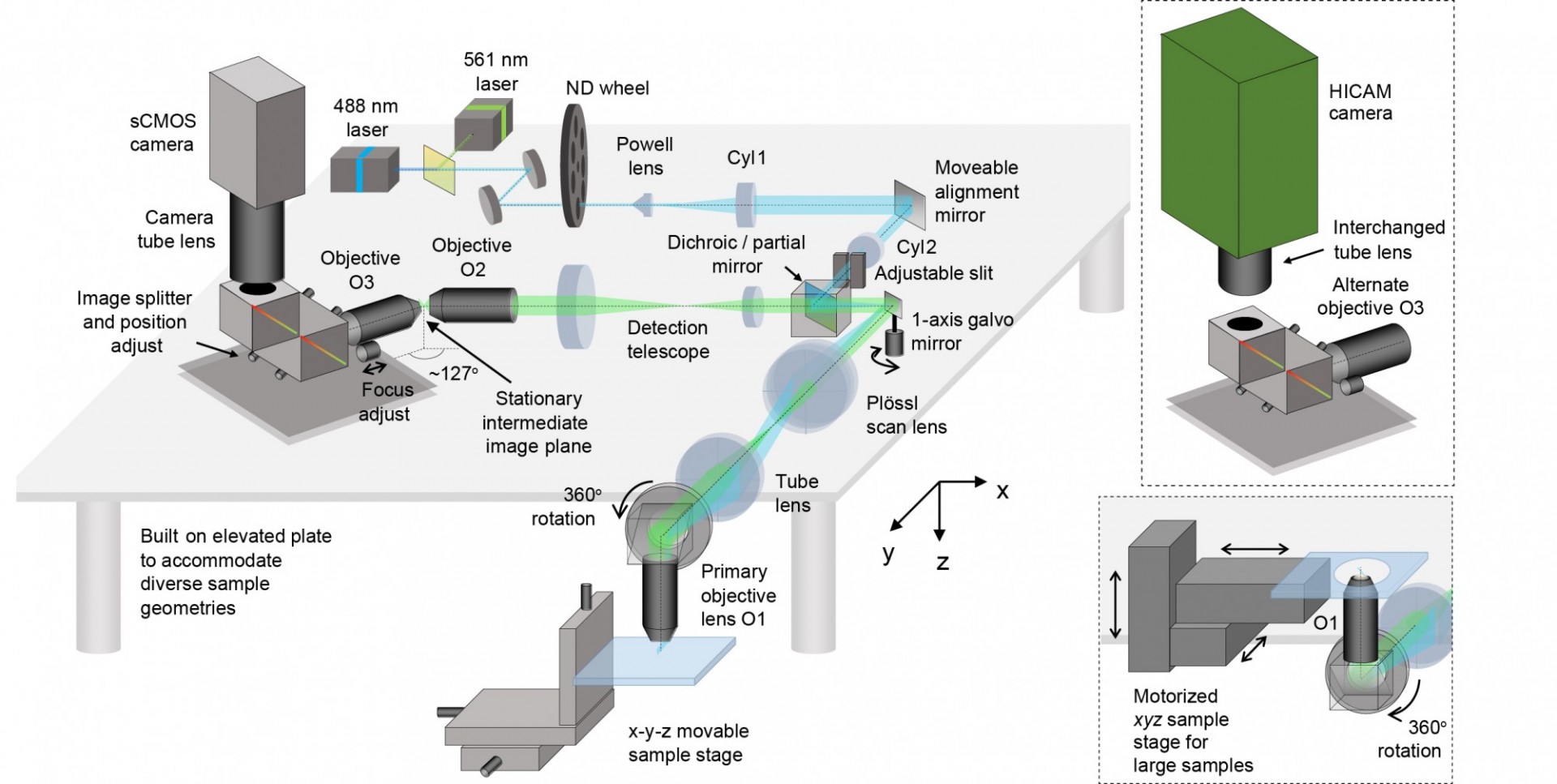
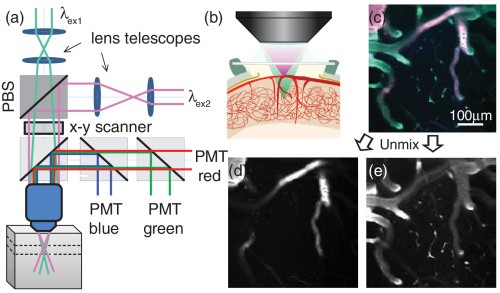
The image above shows the layout of SCAPE 2.0. The system uses a single scanning galvanometer mirror to both move the light sheet back and forth, and to descan returning light to form a stationary oblique image at the intermediate image plane. Objectives O2 and O3 rotate this oblique image so that it is focused onto the high-speed camera. Our image splitter enables two color images to be acquired side by side on the camera chip with no loss of imaging speed. By reading out a reduced number of rows on the camera, we can stream images at up to 18,000 frames per second, yielding imaging at rates of up to 300 volumes per second in the beating zebrafish heart.
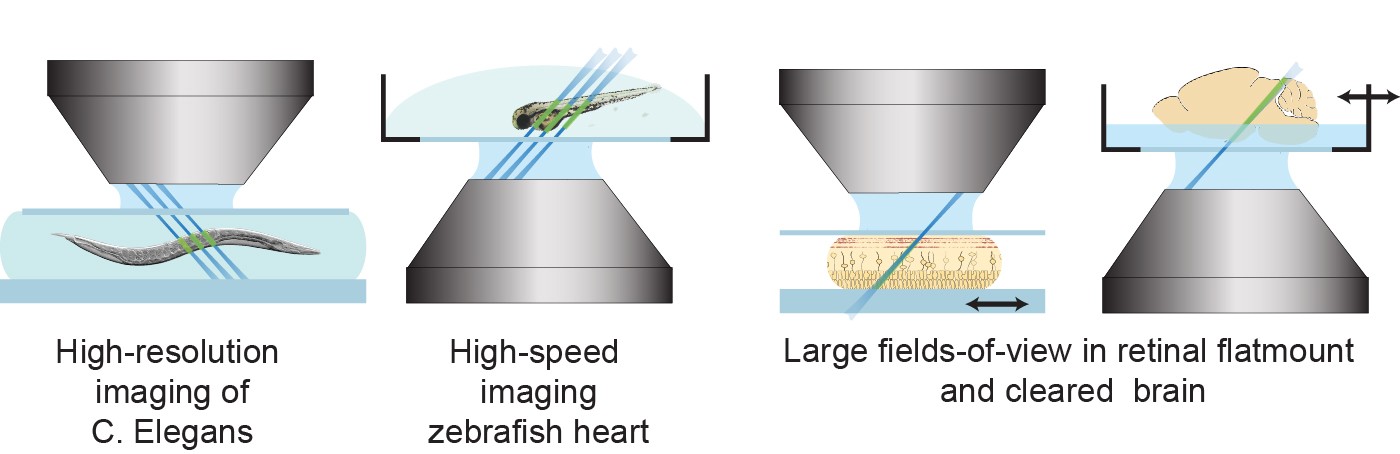
The images below show how SCAPE can be used in many different ways to image a wide range of samples, including C. elegans worms, zebrafish larvae, fixed and flat-mounted specimens and intact tissues.
For imaging results using SCAPE 2.0 see our 'SCAPE applications and Collaborations' pages.
Technology:
Hillman EMC, Voleti V, Li W, Yu H, “Light sheet microscopy in neuroscience”, Annu Rev Neurosci. 42:295-313 (2019).
Hillman EMC, *Voleti V, *Patel K, *Li W, *Yu H, *Perez-Campos C, *Benezra SE, Bruno RM, *Galwaduge PT. “High-speed 3D imaging of cellular activity in the brain using axially-extended beams and light sheets”. Curr Opin Neurobiol. 50:190-200. (2018)
Bouchard MB, Voleti V, Mendes C, Lacefield C, Grueber W, Mann R, Bruno R, Hillman EMC, “Swept, confocally aligned planar excitation (SCAPE) microscopy for ultra-fast, volumetric microscopy of behaving organisms”, Nature Photonics, 9, 113–119 (2015).
Applications:
Xu L, Li W, Voleti V, Hillman EMC, Stuart Firestein, "Widespread Receptor Driven Modulation in Peripheral Olfactory Coding". BioRxiv. 2019 Sep;08.
Vaadia RD†, Li W†, Voleti V, Singhania A, Hillman EMC†, Grueber WB†. "Characterization of Proprioceptive System Dynamics in Behaving Drosophila Larvae Using High-Speed Volumetric Microscopy". Current Biology 29, 935–944 (2019) .†equal contribution .
Nguyen HD, Ullmann JFP, McLachlan GJ, Voleti V, Li W, Hillman EMC, Reutens DC, Janke A, "Whole-volume clustering of time series data from zebrafish brain calcium images via mixture modeling". Stat Anal Data Min: The ASA Data Sci Journal, 2018;11:5–16.
*Open-WFOM coming soon!*
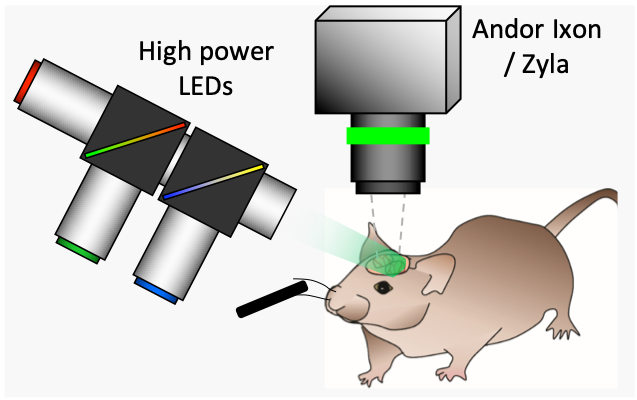
Camera-based spectroscopic optical imaging systems can map functional parameters of living systems by monitoring changes in absorption and/or fluorescence contrast. These systems have been used to study biological questions ranging from cortical hemodynamics to more recent applications in dynamic molecular imaging of pharmacokinetics. We are have developed several high-speed CCD-camera based spectroscopic optical imaging systems that are not hindered by the image acquisition rate limits which have typically constrained spectroscopic imaging systems. This is acheived by free-running the system's camera at its maximum rate, and using its electronic exposure signal to generate a drive waveform for a sequence of different high power, appropriately filtered light emitting diodes (LEDs). Our new high-speed systems are capable of imaging both absorption and fluorescence contrast in parallel at rates exceeding 100 frames per second and greater than 1 megapixel spatial resolution. With appropriate optics, our systems can image living systems ranging in size from small animals to single cells.
So-called 'Optical Intrinsic signal imaging' (OISI) has been used for over 20 years to examine cortical responses to stimulus. In its simplest form, such imaging requires only a light source at a specific wavelength, and a camera to detect changes in the amount of light remitted from the brain's surface. However, it is now widely understood that changes in reflectance of the brain are due to changes in the local concentration of absorbing oxy- and deoxy-hemoglobin. It is therefore possible to make OISI more quantitative if multiple wavelengths of illumination are used, and modeling of light propagation is employed to convert measurements of light intensity into changes in oxy- and deoxy-hemoglobin. A further extension of this kind of imaging is to add fluorescent voltage or calcium sensitive dyes to the brain to allow equivalent imaging of neuronal activity. See Hillman, 2007 and Bouchard et al, 2009 for more details.
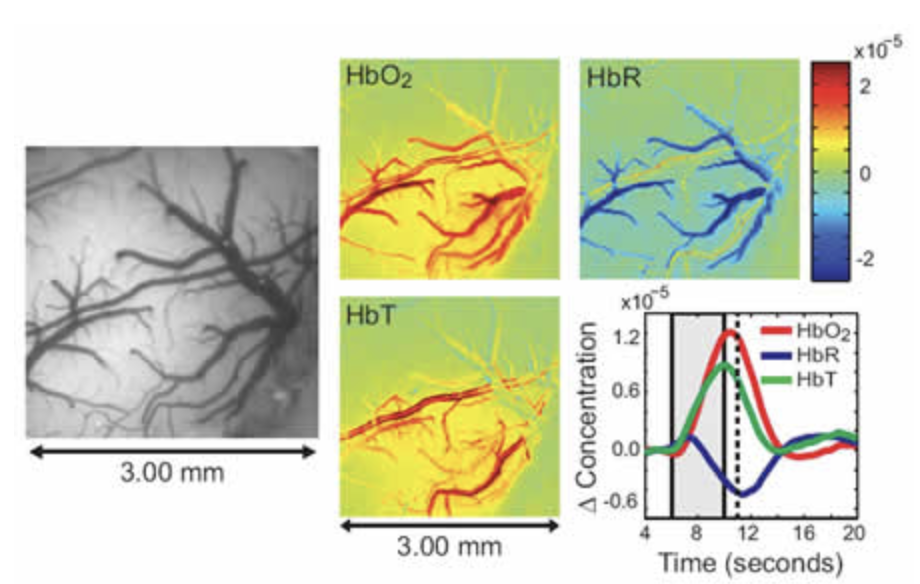
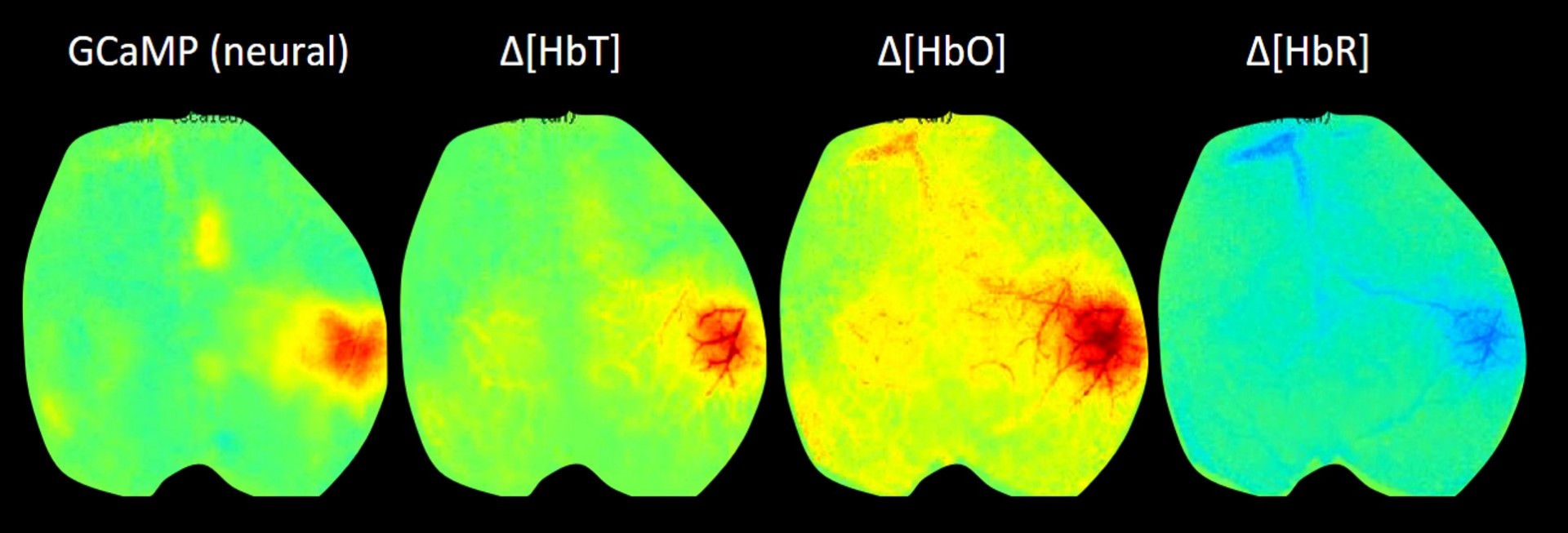
Our latest version of WFOM uses an Andor Zyla 4.2+ camera and up to 4 strobed LEDs to capture both neural activity and hemodynamics in awake, behaving mice. We are using WFOM for a wide range of studies to look at Neurovascular coupling and brain-wide Resting state activity in health and disease. We have also published papers detailing the theoretical basis of conversion of WFOM signals into hemodynamic parameters, and the importance of correctly accounting for cross-talk between hemodynamics and fluorescence signals from calcium indicators such as GCaMP.
Check back soon for our 'Open-WFOM' design.
Technology
Ma Y, Shaik MA, Kim SH, Kozberg MG, Thibodeaux DN, Zhao HT, Yu H., and Hillman EMC, ‘Wide-field optical mapping of neural activity and brain haemodynamics: considerations and novel approaches’. Philos Trans R Soc Lond B Biol Sci. 371 p. 1705 (2016). ** Note CORRECTION (sorry!)
Sun R*, Bouchard MB*, Hillman EMC "SPLASSH: Open source software for camera-based high-speed, multispectral in-vivo optical image acquisition" Biomed Opt Expr, 1 (2), 385-397 (2010). * Equal contribution first authors.
Bouchard MB*, Chen BR*, Burgess SA, Hillman EMC, "Ultra-fast multispectral optical imaging of cortical oxygenation, blood flow and intracellular calcium dynamics", Optics Express, 17 (18), 15670-15678, (2009).* Equal contribution first authors.
Applications:
Montgomery MK, Kim S, Dovas A, Patel K, Mela A, Humala N, Zhao H, Thibodeaux D, Shaik M, Ma Y, Grinband J, Chow D, Schevon C, Hillman EMC, Canoll P, "Glioma-induced alterations in neuronal activity and neurovascular coupling during disease progression". BioRxiv. 2019 Sep;10.
Ma Y, Shaik MA, Kozberg MG, Kim SH, Portes JP, Timerman D, Hillman EMC. "Resting-state hemodynamics are spatiotemporally coupled to synchronized and symmetric neural activity in excitatory neurons." Proceedings of the National Academy of Sciences. PNAS, 113 (52) E8463–E8471 (2016).
Kozberg MG, Ma Y, Kim SH, A. SM, and Hillman EMC, "Rapid postnatal expansion of neural networks occurs in an environment of altered neurovascular and neurometabolic coupling". J Neurosci, , 36(25): 6704-6717 (2016).
Chen BR, Kozberg MG, Bouchard MB, Shaik MA, Hillman EMC, "A critical role for the vascular endothelium in functional neurovascular coupling in the brain", JAHA, 3: e000787, (2014).
Cayce JM, Bouchard MB, Chernov MM, Chen BR, Grosberg LE, Jansen ED, Hillman EMC, Mahadevan-Jansen A. "Calcium imaging of infrared-stimulated activity in rodent brain", Cell Calcium, 55 (4), 183-190(2014).
Rayshubskiy A, Wojtasiewicz TJ, Mikell CB, Bouchard MB, Timerman D, Youngerman BE, McGovern RA, Otten ML, Canoll, PD, McKhann II GM, Hillman,EMC. "Direct, intraoperative observation of ~0.1 Hz hemodynamic oscillations in awake human cortex: Implications for fMRI." NeuroImage, 87, 323-331, (2014).
Chen BR, Bouchard MB, McCaslin AFH, Burgess SA, Hillman EMC, "High-speed vascular dynamics of the hemodynamic response", Neuroimage, 54 (2), 1021-1030 (2011).

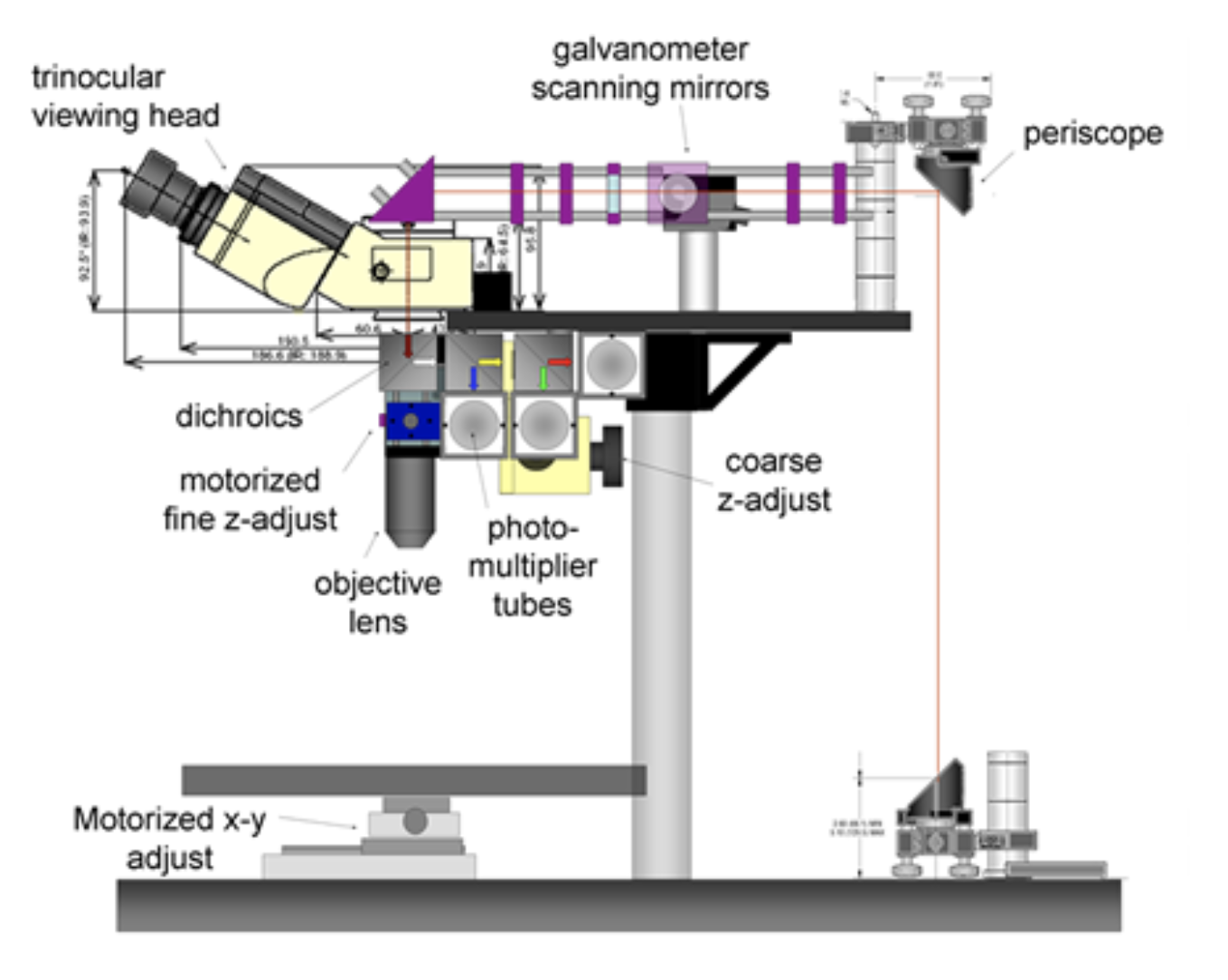
Our two-photon microscope design is optimized for in-vivo imaging. Its three emission channels and flexible filter configuration allow imaging of almost any source of contrast including second harmonic generation, intrinsic fluorescence, fluorescent proteins, active dyes and conventional fluorescent stains. 3 or more sources of contrast can be distinguished and imaged in parallel by the system at up to 30 frames per second, to depths exceeding 500 microns in living tissues. The system is fully automated with all stages and instrument settings controllable via a Matlab graphical user interface (GUI). This software also controls the system's two Spectra Physics MaiTai Ti:Sapphire lasers, allowing simultaneous spectrally-encoded imaging of two planes and synchronous wavelength scanning and imaging allowing hyperspectral microscopy. We have also implemented spatial light modulator-based adaptive optics into the system.
Hyperspectral two-photon microscopy
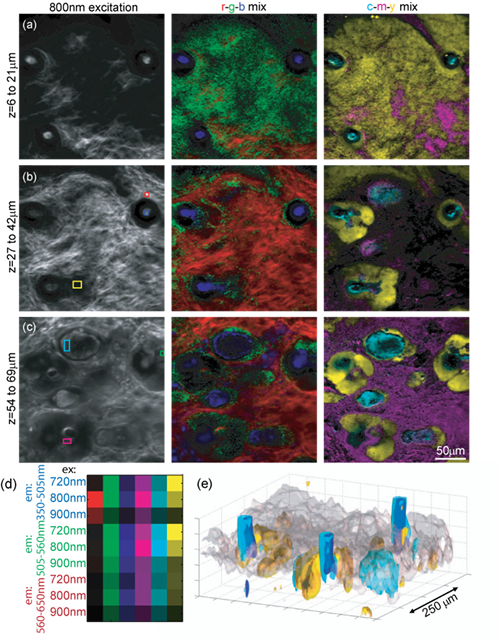
We have constructed a two-photon microscope for high-speed in-vivo imaging. While the system was originally constructed for brain imaging, we have found that the intrinsic fluorophores and sources of second harmonic generation in the skin can allow in-vivo imaging of 3D dermal structures that rival conventional histology. Furthermore, we have developed a novel hyperspectral imaging approach that allows us to clearly delineate the different constituents of skin allowing segmentation and quantitative analysis of different structures.
What is Hyperspectral Two-Photon Microscopy?
Our new technique exploits the tunability of most Ti:Sapphire lasers to acquire detailed spectroscopic information about every voxel in an image. Our system uses a Spectra Physics MaiTai Ti:Sapphire laser which can scan from 710 to 920nm and also incorporates 3 spectrally resolved detectors for collection of emitted light. Therefore, by sequentially acquiring images while scanning the excitation wavelength of our system, we can collect image data in which each pixel is an excitation-emission map of the fluorescent (or SHG-generating) substance in that pixel.
We can take two approaches to analyzing this data: 1) We can use basic spectral unmixing to delineate all of the objects within the image that have the same spectral signature. With intrinsic signal imaging of skin, this means that sebaceous glands can be distinguished from hair follicles, dermal collagen, keratinocytes etc. In stained samples, this would allow rapid segmentation of a large number of dyes. 2) We can fit the excitation / emission spectra of each voxel to the spectra of known constituents of skin, such as collagen SHG, NADH, FAD, keratin, elastin etc. When combined with 3D scanning, we can generate detailed volumetric renderings of living tissue that provide significant advantages over conventional ex-vivo histology. While detailed analysis of intrinsic emission spectra has previously been explored, excitation spectra a much more robust for unmixing. This is because emitted (usually visible) light is strongly attenuated by absorbers in tissue such as hemoglobin, meaning that emission spectra can exhibit depth-dependent distortion of their spectra making unmixing and spectral fitting challenging. Excitation spectra are much more tolerant of this effect because there are few absorbers in tissue between 710 and 920nm, so spectral signatures are better maintained. We are utilizing this technique to study diseased and health models of skin, as well as exploring the potential of hyperspectral two-photon microscopy for imaging other intact tissues.
Multi-plane two-photon using spectral encoding
Upon adding a second Ti:Sapphire laser to our two-photon system, we realized we could do simultaneous dual-plane imaging by using dual-fluorescent labeling and focusing two scanning spots of different wavelengths at different depths. Spectral unmixing was then used to separate out contributions from the two planes.
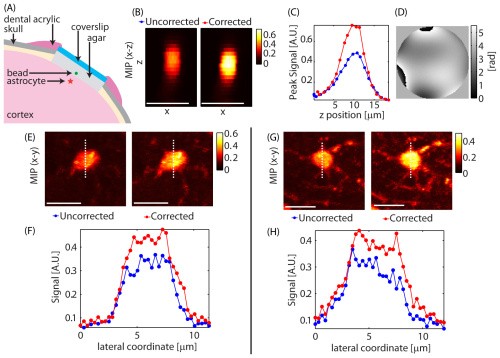
Adaptive optics to improve two-photon in-vivo
Using a spatial light modulator within our beam-path, we performed adaptive optics-based correction of our focal point based on iterative Zernike functions optimized based on two-photon signal. SR101 labeled astrocytes were used as in-vivo guide-stars enabling rapid optimization.
Technology
Galwaduge PT, Kim SH, Grosberg LE, Hillman EMC, "Simple wavefront correction framework for two-photon microscopy of in-vivo brain," Biomed. Opt. Express 6, 2997-3013 (2015)
Grosberg LE, Chen BR, Hillman EMC, "Simultaneous multi-plane in-vivo non-linear microscopy using spectral encoding", Optics Letters, 37(14), 2967-2969 (2012).
Grosberg LE, Radosevich AJ, Asfaha S, Yang X, Wang TJ, Hillman EMC, "Spectral characterization and unmixing of intrinsic contrast in intact normal and diseased gastric tissues using hyperspectral two-photon microscopy" PlosOne, 6(5): e19925 (2011).
Radosevich AJ, Bouchard MB, Burgess SA, Chen BR, Hillman EMC. “Hyper-spectral in-vivo two-photon microscopy of intrinsic fluorophores”, Optics Letters, 33 (18), 2164-2166, (2008).
Applications
Cayce JM, Bouchard MB, Chernov MM, Chen BR, Grosberg LE, Jansen ED, Hillman EMC, Mahadevan-Jansen A. "Calcium imaging of infrared-stimulated activity in rodent brain", Cell Calcium, 55 (4), 183-190 (2014).
McCaslin AFH, Chen BR, Radosevich AJ, Cauli B, Hillman EMC, "In-vivo 3D morphology of astrocyte-vasculature interactions in the somatosensory cortex: implications for neurovascular coupling", J Cereb
Blood Flow Metab, 31, 795-806 (2011).
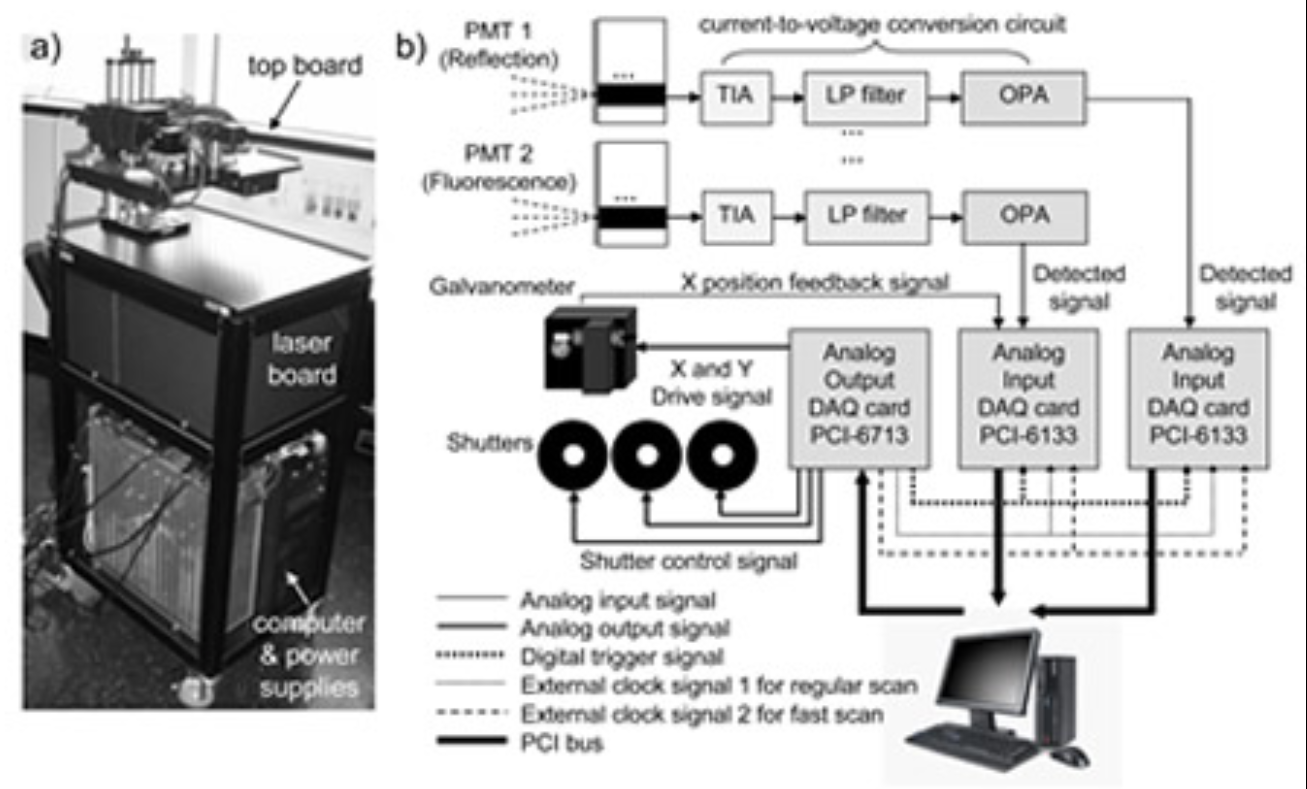
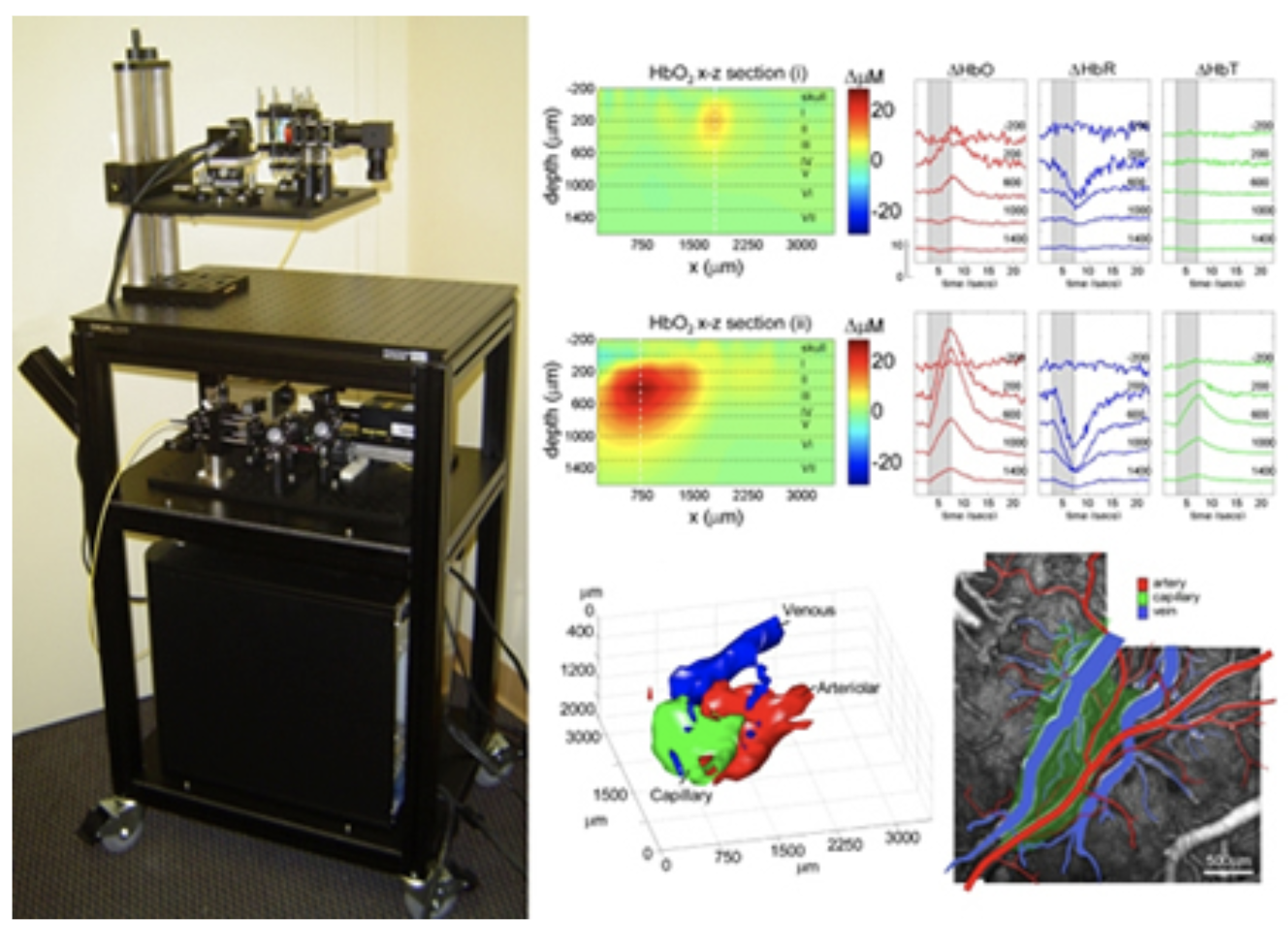
LOT uses laser-scanning instrumentation similar to a confocal microscope. Light is serially injected into the surface of a tissue. Once in tissue, light will undergo scattering events, encountering absorbers and fluorophores along its path. Some of this backscattered light will be remitted from the tissue surface at different distances from the source position. Light emitting further from the incident source has, on average, travelled more deeply into the tissue. LOT therefore acquires depth sensitive information by measuring off axis backscattered light. Whereas a confocal microscope rejects off axis backscattered light with a pinhole, LOT replaces the pinhole with a slit and uses an array of optical detectors to acquire depth sensitive tomographic measurements. The three-dimensional distribution of optical contrast represented by these measurements can be deduced using models of light propagation, and a tomographic style image reconstruction.
We have applied LOT to a range of living tissues including the brain, heart and skin. For more information on applications of LOT see Hillman et al 2007, Hillman et al 2007b, Yuan et al, 2009, Hillman and Burgess 2009, Burgess and Hillman 2010 and Burgess et al 2010.
Laminar Optical Tomography can add a depth-dimension to conventional multi-spectral optical brain imaging. LOT can currently image hemodynamic activity to > 2mm, with 100-200 micron resolution at over 80 frames per second.
To date, we have demonstrated that LOT can resolve the oxy– and deoxy-hemoglobin responses in individual vascular compartments in the somatosensory cortex during forepaw stimulation. This was achieved using a unique spatiotemporal linear fitting procedure based on the characteristic temporal behavior of the arteriolar, capillary and venous responses. We have now added fluorescence imaging capabilities to LOT to allow simultaneous depth-resolved imaging of voltage or calcium sensitive fluorescent dyes to enable 3D studies of neurovascular coupling in the living brain through the full thickness of the cortex. See Hillman et al, 2007 for more details.
Technology
Burgess SA, Ratner D, Chen BR, Hillman EMC, "Fiber-Optic and Articulating Arm Implementations of Laminar Optical Tomography for Clinical Applications" Biomed Opt Expr, 1 (3), 780-790, (2010).
Burgess SA and Hillman EMC, "Laminar Optical Tomography", Chapter in 'Handbook of Biomedical Optics'. Eds: Boas DA, Pitris C, Ramanujam N (Taylor & Francis) (2010)
Yuan B, Burgess SA, Iranmahboob AK, Bouchard MB, Lehrer N, Bordier C, Hillman EMC, "System for rapid, simultaneous, high resolution, depth-resolved fluorescence and absorption optical imaging", Rev. Sci. Instrum. 80, 043706-1 (2009).
Hillman EMC, Burgess SA. "Sub-millimeter Resolution 3D Optical Imaging of Living Tissue using Laminar Optical Tomography", Laser & Photonics Reviews, 3 (1-2), 159-180, (2009).
Burgess SA, Bouchard MB, Yuan B, Hillman EMC. “Simultaneous Multi-Wavelength Laminar Optical Tomography"Optics Letters, 33 (22), 2710-2712 (2008). *Also selected to appear in the Virtual Journal of Biomedical Optics, 4 (1), 2710.
Hillman EMC, Devor A, Bouchard M. B, Dunn A. K, Krauss GW, Skoch J, Bacskai J, Dale A. M, Boas D. A. “Depth-resolved Optical Imaging and Microscopy of Vascular Compartment Dynamics During Somatosensory Stimulation”, NeuroImage, 35(1): 89-104 (2007)
Hillman, E. M. C, Boas, D. A, Dale, A. M, Dunn, A. K, "Laminar Optical Tomography: demonstration of millimeter-scale depth-resolved imaging in turbid media", Optics Letters, 29, (14), 1650-1652 (2004).
Applications
Muldoon, TJ, Burgess SA, Chen BR, Ratner D, and EMC. Hillman, "Analysis of skin lesions using laminar optical tomography". Biomed. Opt. Express, 3(7): p. 1701-1712 (2012).
Hillman EMC, Bernus O, Pease E, Bouchard MB. Pertsov A. “Depth-resolved optical imaging of transmural electrical propagation in perfused heart”, Optics Express, 15 (26), 17827-17841, 2007
Hillman EMC, Devor A, Bouchard M. B, Dunn A. K, Krauss GW, Skoch J, Bacskai J, Dale A. M, Boas D. A. “Depth-resolved Optical Imaging and Microscopy of Vascular Compartment Dynamics During Somatosensory Stimulation”, NeuroImage, 35(1): 89-104 (2007)
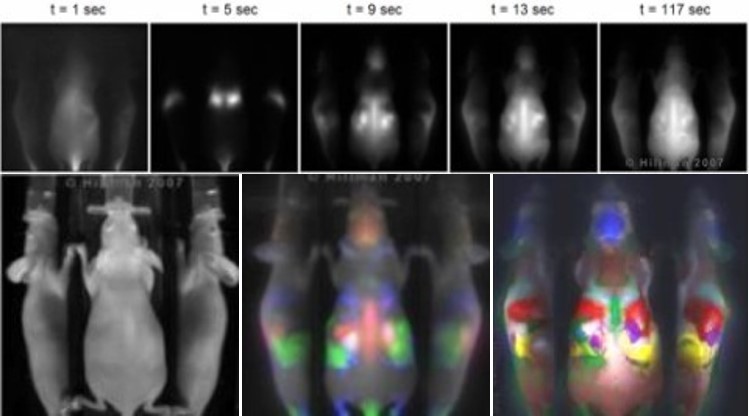
Dynamic Contrast-Enhanced (DyCE) imaging is a way to acquire detailed images of small animals using optical methods. This work was published in Nature Photonics in 2007, and licensed to CRi and then PerkinElmer.
Conventional molecular imaging uses targeted dyes to reveal in-vivo biochemical processes such as those specific to cancer tumors and is used by researchers and pharmaceutical companies for drug development and disease process research. Developed dyes may ultimately also be useful for clinical diagnostics and therapies.
However, small animal imaging can be difficult to interpret because of poor resolution, specificity and difficulties in interpreting images. Our new technique addresses the need for improved imaging resolution, dye specificity, contrast and anatomical co-registration for in-vivo imaging in small animals.
We found that the in-vivo dynamics of a dye can be harnessed to reveal incredible detail, clearly delineating internal organs. Previous attempts to provide anatomical information for optical molecular imaging included incorporation of x-ray CT, x-ray and MRI modalities. Our method offers improved performance, versatility and co-registration at a fraction of the cost and complexity of multi-modality systems.

Technology
Hillman EMC, Amoozegar CB, Wang T, McCaslin AFH, Bouchard MB, Mansfield JR, Levenson RM, “In-vivo optical imaging and dynamic contrast methods for biomedical research” Phil. Trans. Royal Soc A. 369 (1955), 4620-4643 (2011).
Hillman EMC and Moore A. "All-optical anatomical co-registration for molecular imaging of small animals using dynamic contrast", Nature Photonics, 1 (9), 526 - 530, 2007.
Applications
Miller J, Wang ST, Orukari I, Prior J, Sudlow G, Su X, Liang K, Tang R, Hillman EMC, Weilbaecher KN, Culver JP, Berezin MY, Achilefu S. “Perfusion-based fluorescence imaging method delineates diverse organs and identifies multifocal tumors using generic near-infrared molecular probes.”, J Biophotonics. Apr;11(4):e201700232 (2018).
Amoozegar CB, Wang T, Bouchard MB, McCaslin AFH, Blaner WS, Levenson RM, Hillman EMC, "Dynamic contrast enhanced optical imaging of in-vivo organ function", J Biomed Opt, 17(9), 096003 (2012)